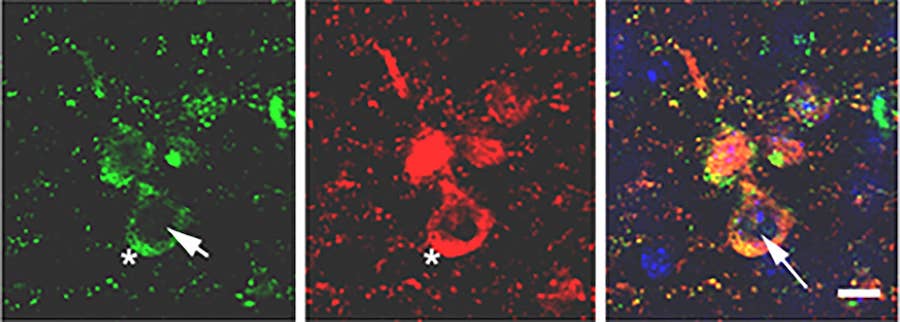
Regulatory proteins and epigenetic mechanisms including DNA methylation and histone modifications tightly control gene expression. Increasing evidence supports the idea that DNA methylation is dynamically regulated in post mitotic neurons with crucial functions in memory and synaptic plasticity. Our earliest findings showed aberrant localization of the methylation enzyme DNMT1 in the cytoplasm of neuronal cells, as a result of its interaction with aggregated alpha-synuclein, the protein accumulated in Lewy Bodies and the pathological hallmark of Parkinson’s disease (PD). This abnormal localization was associated with global and gene specific changes in DNA methylation.
Our subsequent study identifying concordant methylation changes in brain and blood from PD cases (Epigenetics 2013 8(10): 1030) encouraged the exploration of methylation signatures as potential biomarkers for PD diagnosis and/or to monitor disease-modifying therapies.
We continue expanding our research by profiling larger cohorts of PD patients and control subjects, including longitudinal blood samples from the Harvard Biomarkers Study (HBS) and the Parkinson’s Progression Markers Initiative (PPMI). This effort, supported by generous grants from the Michael J Fox Foundation, has already identified multiple pathways affected by epigenetic alterations that may contribute to disease progression, pathology and associated with response to dopamine replacement therapy.
