Multiple system atrophy (MSA) is a fatal synucleinopathy characterized by the accumulation of α-synuclein aggregates in oligodendroglia, leading to demyelination and neurodegeneration. My lab is working to decipher the interaction between α-synuclein misfolding/accumulation and epigenetic regulation. In particular, we are investigating the involvement of microRNAs (miRNAs) in MSA as major regulators of the transcriptome, as well as the potential of manipulating miRNA levels to act as a therapeutic tool.
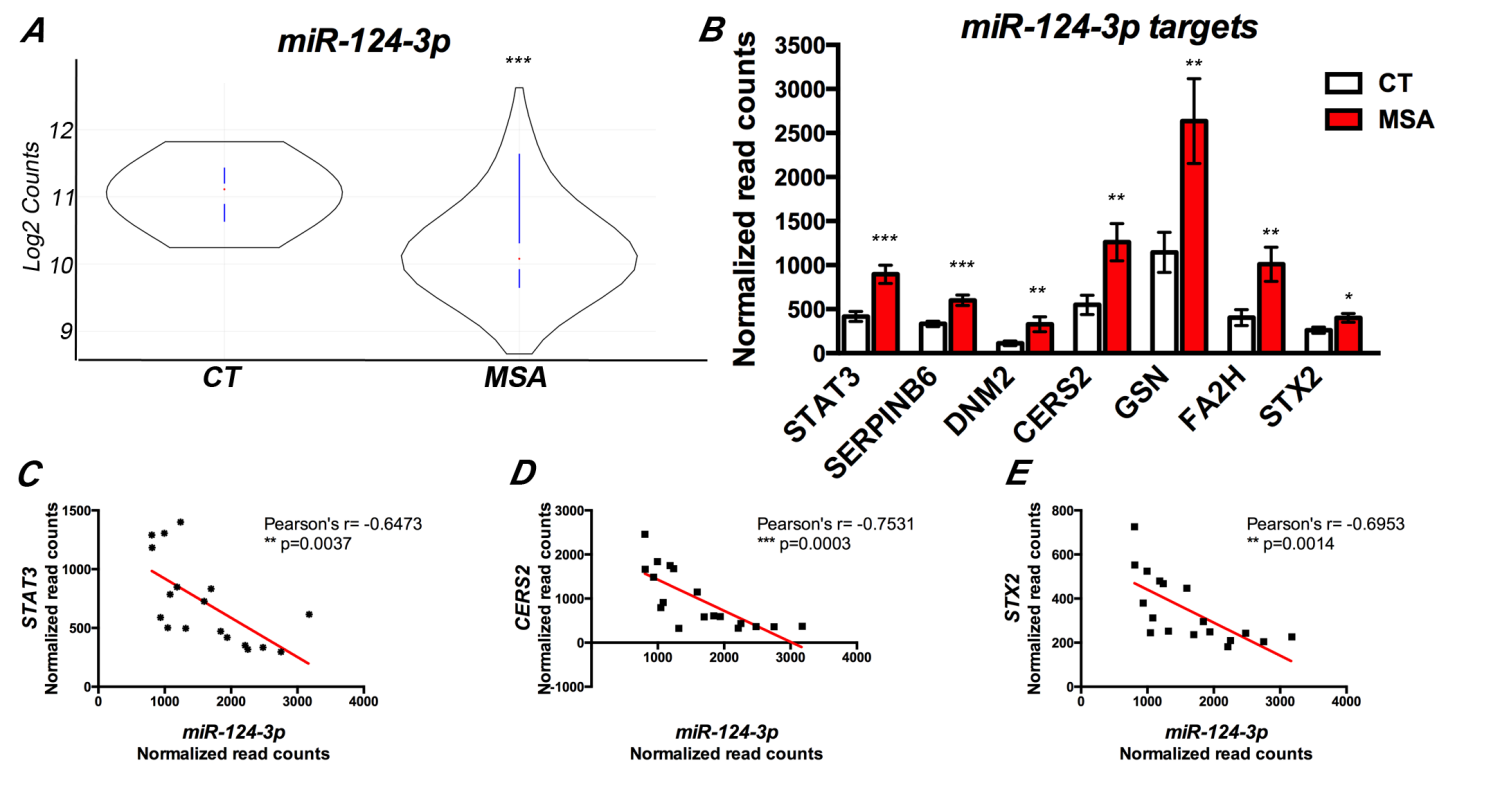
Alterations in specific miRNAs have been reported in brain tissue, serum, and cerebrospinal fluid from MSA patients. Still, a causal link between deregulation of miRNA networks and pathological changes in the transcriptome remained elusive. We profiled ~800 miRNAs in the striatum of MSA patients, in combination with screening of 700 transcripts associated with neurodegeneration, to determine the impact of miRNA deregulation on the transcriptome (Kim et al. Mol. Neurobiol. 2019). We identified 60 miRNAs involved in extracellular matrix receptor interactions, prion disease, inflammation, ubiquitin-mediated proteolysis, and addiction pathways that showed abnormal levels in MSA brains. Correlation of miRNA expression with the abundance of their known targets identified miR-124-3p, miR-19a-3p, miR-27b-3p and miR-29c-3p as key regulators altered in MSA, mainly contributing to neuroinflammation. Finally, our study also uncovered a potential link between Alzheimer’s disease (AD) and MSA pathologies, involving miRNAs and deregulation of BACE1.